Explore clustering results
Initiate the ClusterAnalysis object
ClusTCR provides a number of options for exploration of clustering results. To perform these analysis, you must initiate a ClusterAnalysis object, which takes cluster features as its only argument.
from clustcr import ClusterAnalysis
analysis = ClusterAnalysis(features)
PCA
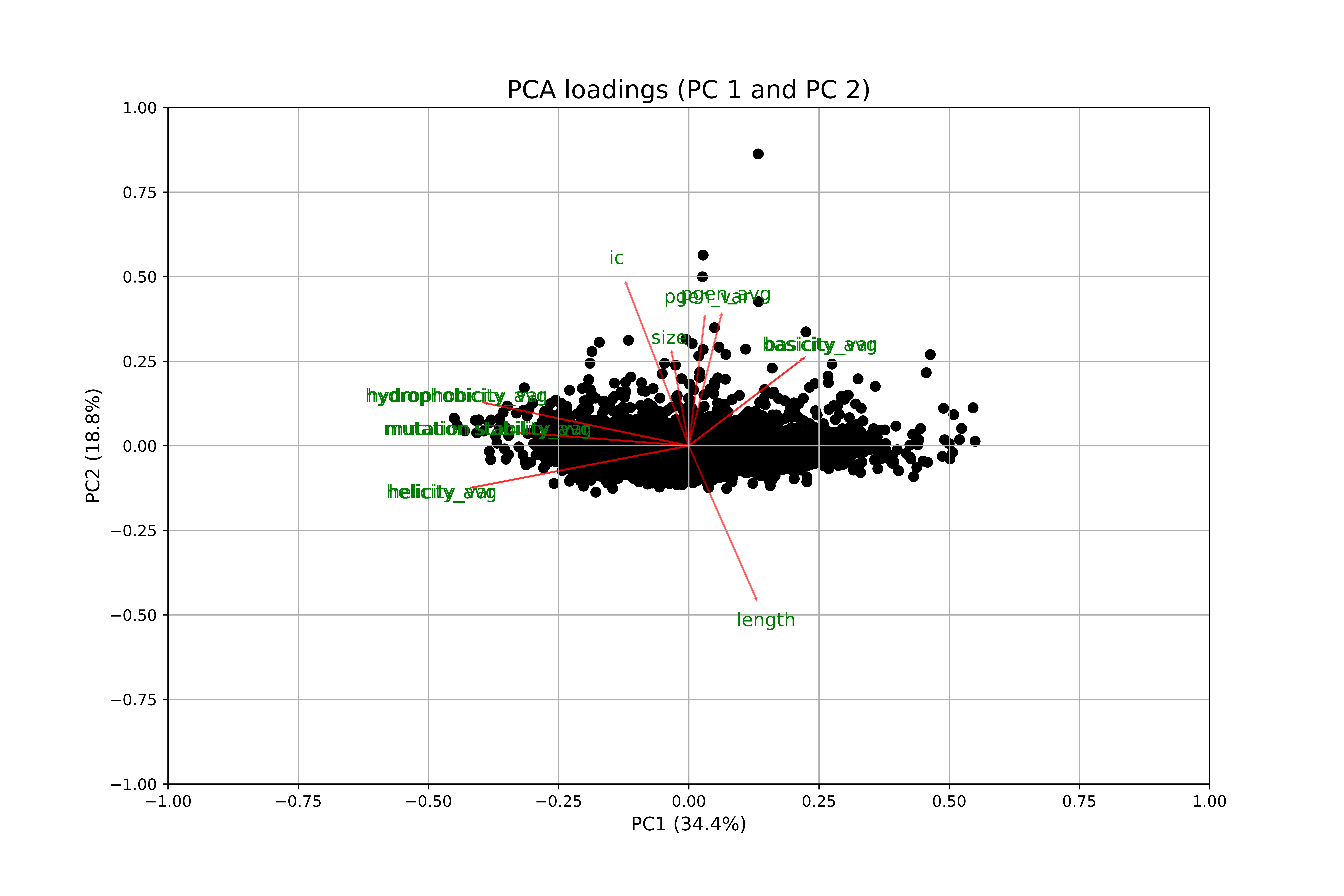
To get a quick overview of the newly generated features, you can perform a principal component analysis (PCA). clusTCR contains a built-in PCA functionality, which can be conveniently executed by performing the .pca() method on a ClusterAnalysis object.
analysis.pca()
Performing a PCA with clusTCR will provide a figure of the PCA loadings.
Analyzing cluster quality
An additional feature of ClusTCR’s analysis module is predicting the quality of a cluster. Here, quality is determined as the purity of a cluster. This feature is particularly useful when no information is available about the target epitopes of the clustered TCR sequences. We trained a classification model that predicts whether an individual cluster will be of good (1) or bad (0) quality. Good clusters have a predicted purity of > 0.90, low quality clusters have a purity < 0.90.
Using the pre-trained model
predictions = analysis.predict_quality()
Training your own model
You can also train your own model by creating a ModelTraining object. To do this, you will need to provide the following three arguments: features (see features page), results from the clustering and epitope data corresponding to the CDR3 sequences used for clustering.
from clustcr import ModelTraining
model = ModelTraining(features, clusters, epitopes)
clf = model.fit()
You can evaluate your own model using the .evaluate() method. This will perform 10-fold stratified cross-validation and outputs a receiver operating characteristic (ROC) curve with corresponding area under the curve (AUC) value.
# Evaluate your own model
model.evaluate()

Save your model using the following method:
model.save(clf, '/path_to_model/my_custom_model.pkl')
If you want to use this model to make quality predictions, just specify the path to your model in the model parameter of the predict_quality() method.